| size | 16841 |
| size_real | 16841 |
| ecount | 2180097 |
| supercluster | 1 |
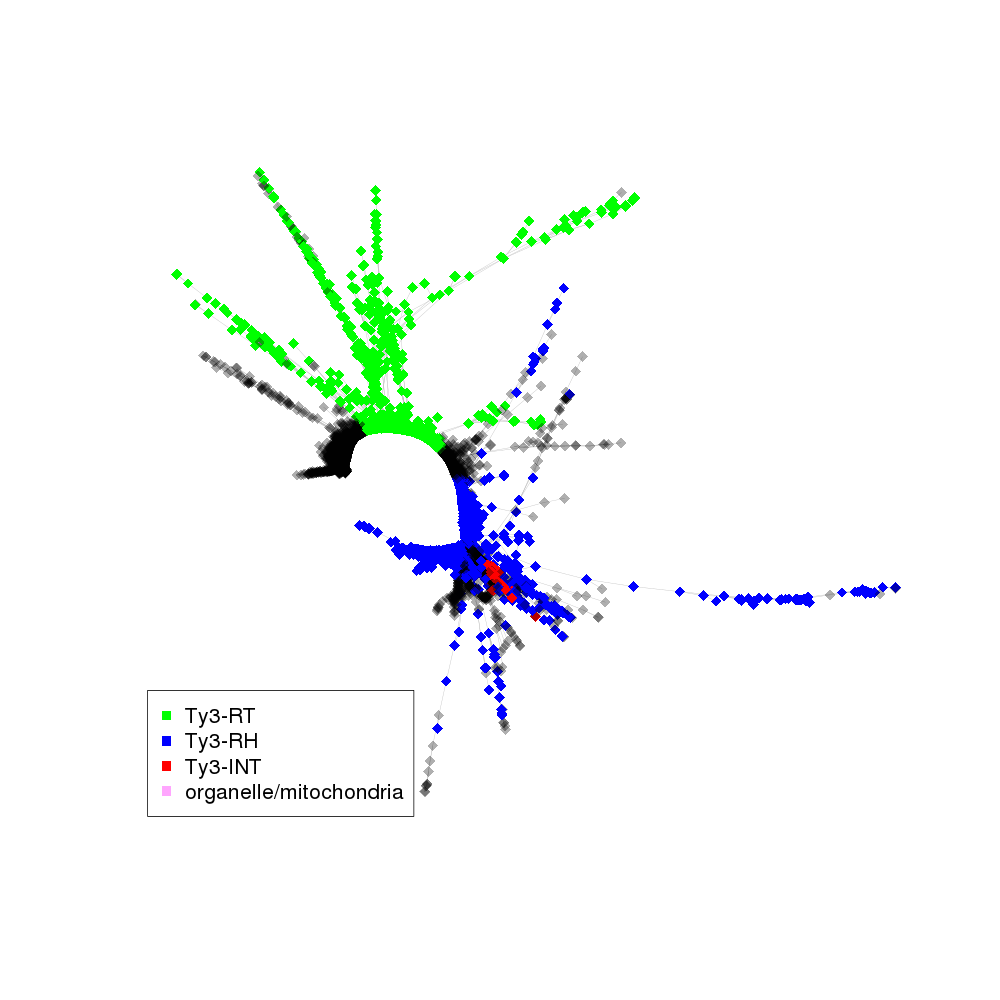
| annotations_summary | 26.77% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIII:Ty3-RT 23.11% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIII:Ty3-RH 3.86% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIII:Ty3-INT 0.33% organelle/mitochondria 0.02% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatII:Ty3-RT 0.02% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Athila:Ty3-RT 0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIV_Ogre:Ty3-RT |
| pair_completeness | 0.783815273805741 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.00142509352176237 |
| satellite_probability | 3.89552186042479e-23 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |
protein domains:

|
|
||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|