| size | 16530 |
| size_real | 16530 |
| ecount | 3571035 |
| supercluster | 1 |
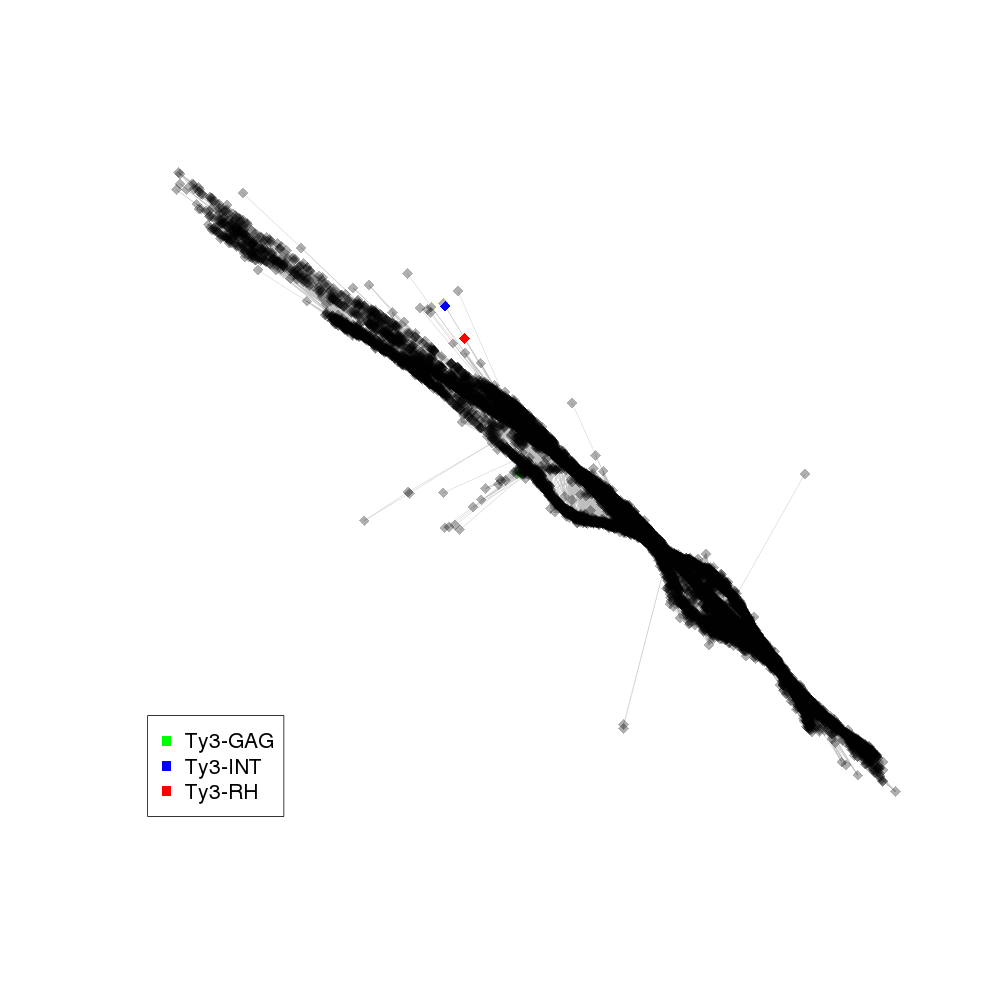
| annotations_summary | 0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Tekay:Ty3-GAG 0.01% Class_I/LTR/Ty3_gypsy/chromovirus/CRM:Ty3-RH 0.01% Class_I/LTR/Ty3_gypsy/chromovirus/Reina:Ty3-INT |
| pair_completeness | 0.740916271721959 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.0126452081316554 |
| satellite_probability | 8.58421306693902e-22 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 0.999999719987332 |
protein domains:

|
|
||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|