| size | 13990 |
| size_real | 13990 |
| ecount | 3035158 |
| supercluster | 32 |
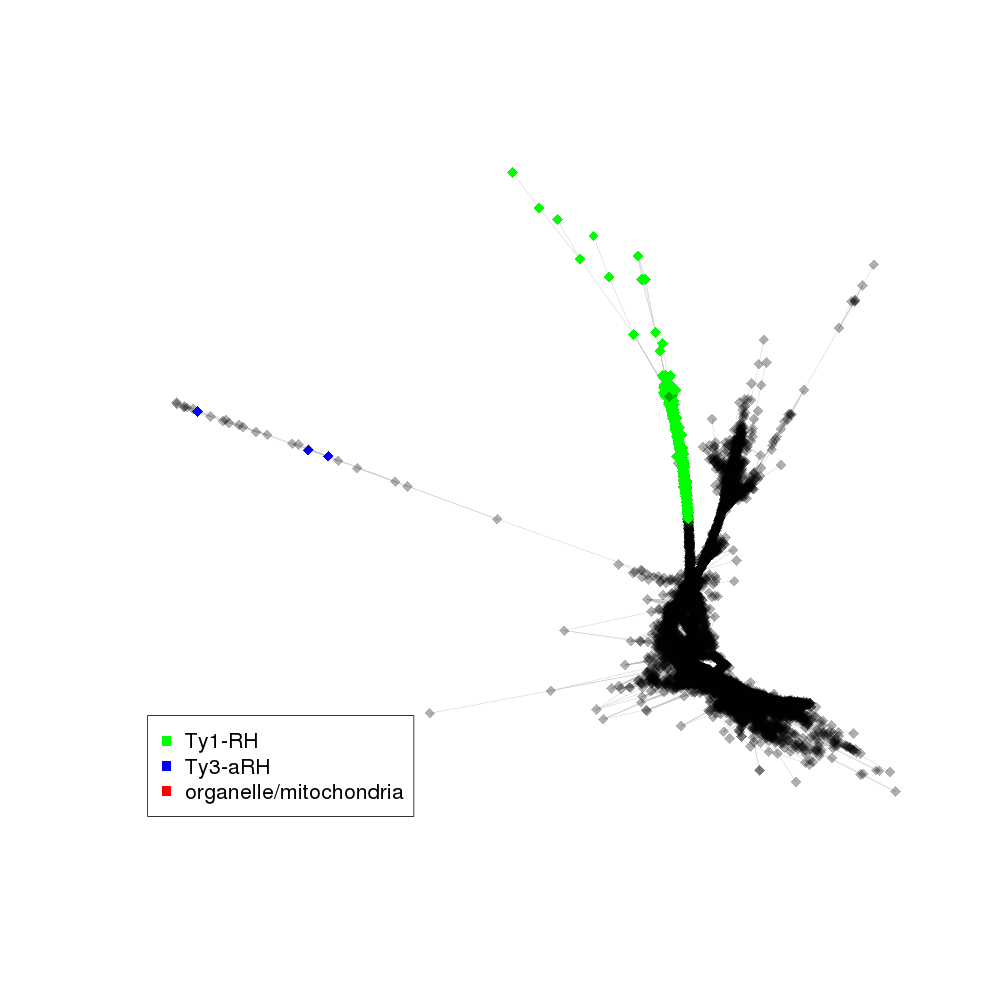
| annotations_summary | 5.63% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-RH 0.02% organelle/mitochondria 0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-aRH 0.01% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatIV_Ogre:Ty3-aRH |
| pair_completeness | 0.642982971227246 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.0129378127233738 |
| satellite_probability | 2.63699184809533e-21 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |
protein domains:

|
|
||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|