| size | 11007 |
| size_real | 11007 |
| ecount | 2781703 |
| supercluster | 16 |
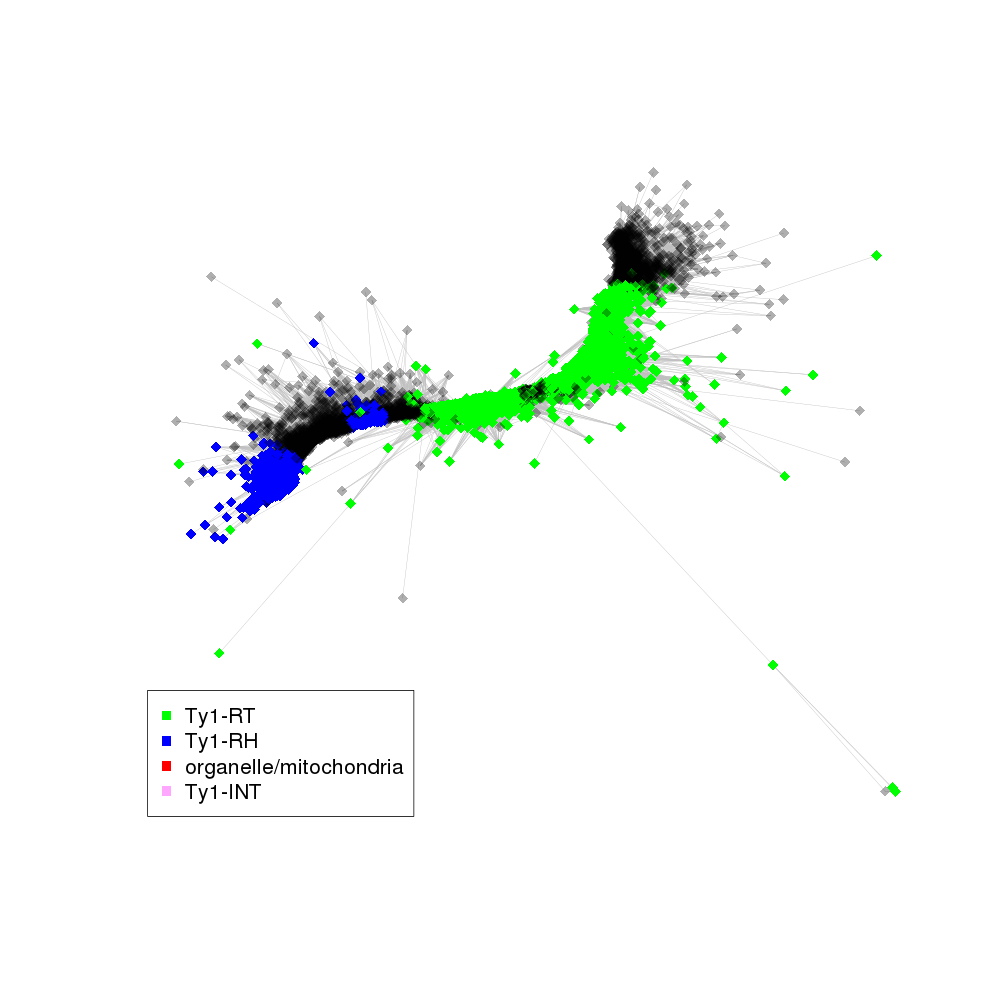
| annotations_summary | 24.03% Class_I/LTR/Ty1_copia/Ale:Ty1-RT 19.55% Class_I/LTR/Ty1_copia/Ivana:Ty1-RT 7.27% Class_I/LTR/Ty1_copia/Ivana:Ty1-RH 3.68% Class_I/LTR/Ty1_copia/Bryco:Ty1-RT 2.96% organelle/mitochondria 2.66% Class_I/LTR/Ty1_copia/Tork:Ty1-RT 2.53% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-RT 1.21% Class_I/LTR/Ty1_copia/Ikeros:Ty1-RT 0.98% Class_I/LTR/Ty1_copia/Ale:Ty1-RH 0.97% Class_I/LTR/Ty1_copia/Gymco-I:Ty1-RT 0.88% Class_I/LTR/Ty1_copia/SIRE:Ty1-RT 0.79% Class_I/LTR/Ty1_copia/Bryco:Ty1-RH 0.36% Class_I/LTR/Ty1_copia/Alesia:Ty1-RH 0.23% Class_I/LTR/Ty1_copia/TAR:Ty1-RT 0.13% Class_I/LTR/Ty1_copia/Tork:Ty1-RH 0.06% Class_I/LTR/Ty1_copia/Angela:Ty1-RT 0.05% Class_I/LTR/Ty1_copia/Gymco-I:Ty1-RH 0.04% Class_I/LTR/Ty1_copia/Ikeros:Ty1-RH 0.03% Class_I/LTR/Ty1_copia/SIRE:Ty1-RH 0.03% Class_I/LTR/Ty1_copia/Ty1-outgroup:Ty1-RH 0.03% Class_I/LTR/Ty1_copia/Angela:Ty1-RH 0.02% Class_I/LTR/Ty1_copia/TAR:Ty1-RH 0.02% Class_I/LTR/Ty1_copia/Ty1-outgroup:Ty1-RT 0.02% Class_I/LTR/Ty1_copia/Osser:Ty1-RT 0.01% Class_I/LTR/Ty1_copia/Osser:Ty1-RH 0.01% Class_I/LTR/Ty1_copia/Ty1-outgroup:Ty1-INT |
| pair_completeness | 0.765640038498556 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.00236213318797129 |
| satellite_probability | 4.71413703996316e-23 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |
protein domains:

|
|
||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|