| size | 8311 |
| size_real | 8311 |
| ecount | 55466 |
| supercluster | 80 |
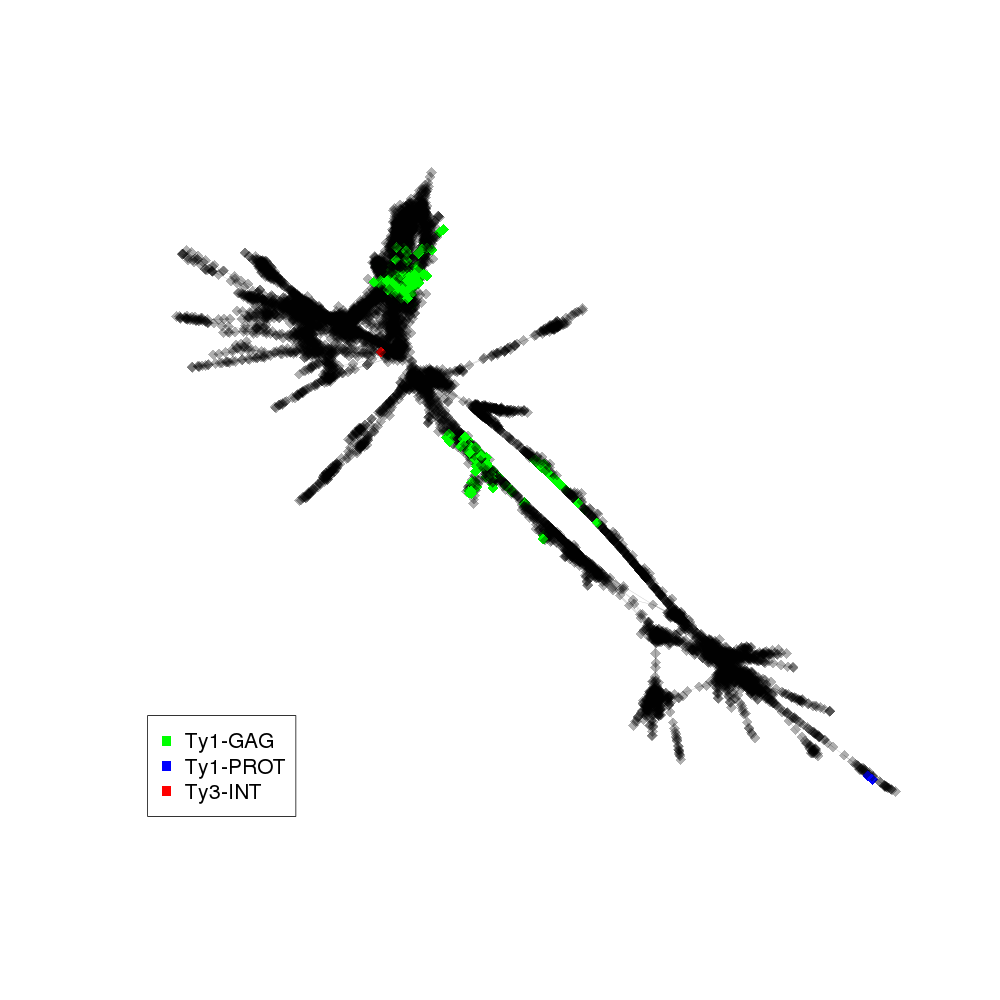
| annotations_summary | 6.04% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-GAG 0.67% Class_I/LTR/Ty1_copia/Tork:Ty1-GAG 0.24% Class_I/LTR/Ty1_copia/Ale:Ty1-GAG 0.14% Class_I/LTR/Ty1_copia/Ivana:Ty1-GAG 0.04% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-PROT 0.01% Class_I/LTR/Ty1_copia/Gymco-I:Ty1-GAG 0.01% Class_I/LTR/Ty3_gypsy/chromovirus/chromo-unclass:Ty3-INT |
| pair_completeness | 0.563981934512608 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.0170857899169775 |
| satellite_probability | 7.64480299365326e-20 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |
protein domains:

|
|
||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|