| size | 7043 |
| size_real | 7043 |
| ecount | 153891 |
| supercluster | 92 |
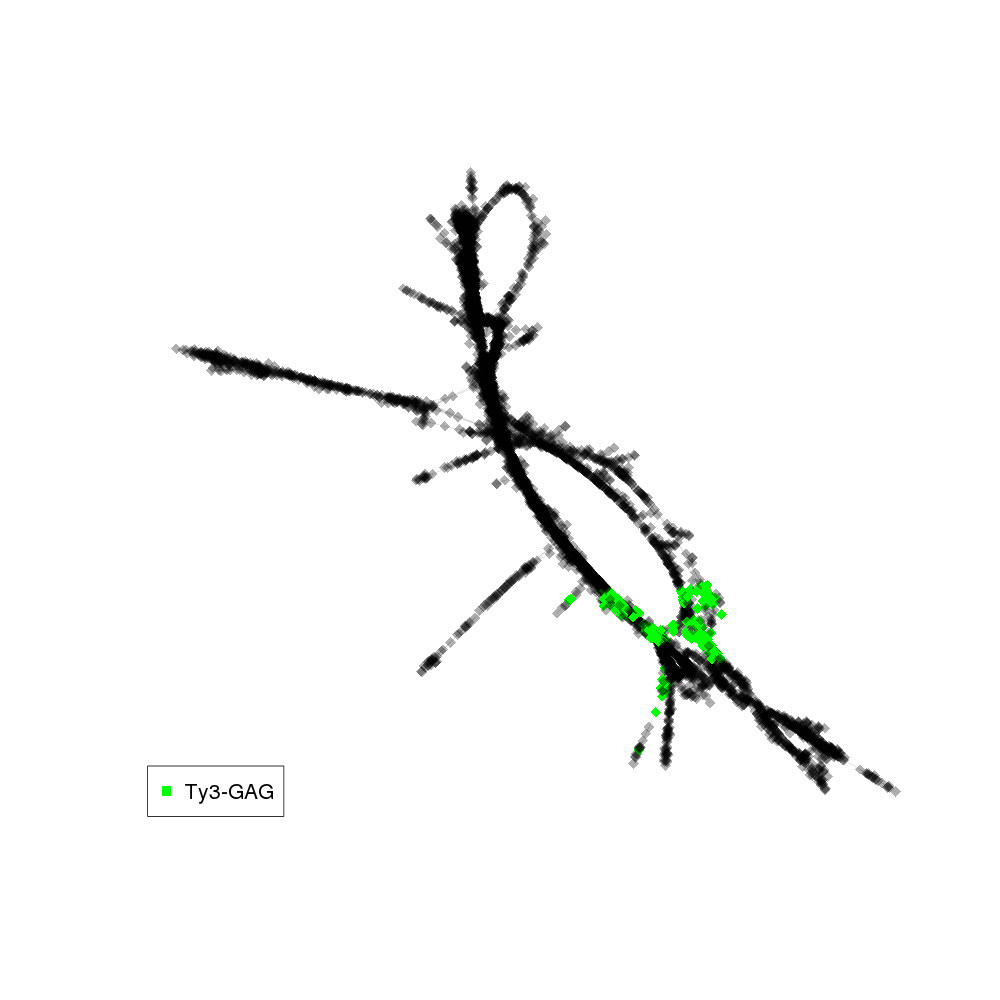
| annotations_summary | 3.38% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatII:Ty3-GAG 1.66% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Ogre_Tat/TatV:Ty3-GAG 0.81% Class_I/LTR/Ty3_gypsy/non-chromovirus/OTA/Athila:Ty3-GAG |
| pair_completeness | 0.79622545269064 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.00340763879028823 |
| satellite_probability | 2.63652108321905e-23 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |
protein domains:

|
|
||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|