| size | 4254 |
| size_real | 4254 |
| ecount | 56863 |
| supercluster | 118 |
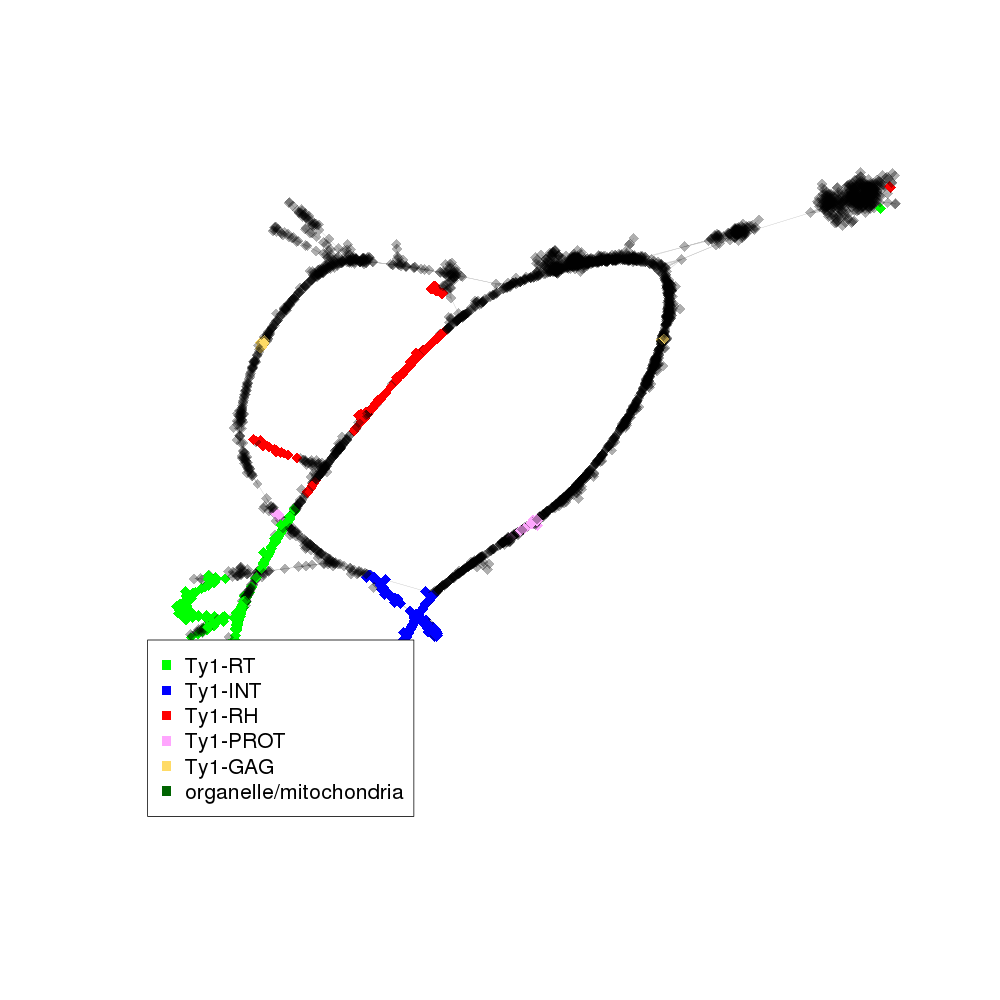
| annotations_summary | 5.27% Class_I/LTR/Ty1_copia/Ale:Ty1-RT 5.10% Class_I/LTR/Ty1_copia/Ivana:Ty1-INT 2.35% Class_I/LTR/Ty1_copia/Ivana:Ty1-RT 2.16% Class_I/LTR/Ty1_copia/Ale:Ty1-RH 1.46% Class_I/LTR/Ty1_copia/Ivana:Ty1-RH 0.87% Class_I/LTR/Ty1_copia/Alesia:Ty1-INT 0.87% Class_I/LTR/Ty1_copia/Tork:Ty1-RT 0.66% Class_I/LTR/Ty1_copia/SIRE:Ty1-INT 0.61% Class_I/LTR/Ty1_copia/Ale:Ty1-INT 0.61% Class_I/LTR/Ty1_copia/Tork:Ty1-INT 0.47% Class_I/LTR/Ty1_copia/Bryco:Ty1-RT 0.45% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-INT 0.35% Class_I/LTR/Ty1_copia/Ivana:Ty1-PROT 0.28% Class_I/LTR/Ty1_copia/Tork:Ty1-PROT 0.26% Class_I/LTR/Ty1_copia/Ale:Ty1-GAG 0.26% Class_I/LTR/Ty1_copia/Ale:Ty1-PROT 0.24% organelle/mitochondria 0.19% Class_I/LTR/Ty1_copia/Ty1-outgroup:Ty1-INT 0.16% Class_I/LTR/Ty1_copia/SIRE:Ty1-RH 0.16% Class_I/LTR/Ty1_copia/Angela:Ty1-RT 0.16% Class_I/LTR/Ty1_copia/Ikeros:Ty1-INT 0.12% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-RT 0.09% Class_I/LTR/Ty1_copia/TAR:Ty1-INT 0.09% Class_I/LTR/Ty1_copia/Gymco-II:Ty1-RH 0.07% Class_I/LTR/Ty1_copia/Gymco-I:Ty1-RT 0.05% Class_I/LTR/Ty1_copia/Ty1-outgroup:Ty1-GAG 0.05% Class_I/LTR/Ty1_copia/Alesia:Ty1-GAG 0.05% Class_I/LTR/Ty1_copia/Ty1-outgroup:Ty1-PROT 0.05% Class_I/LTR/Ty1_copia/Ikeros:Ty1-RT 0.02% Class_I/LTR/Ty1_copia/SIRE:Ty1-GAG 0.02% Class_I/LTR/Ty1_copia/Osser:Ty1-INT 0.02% Class_I/LTR/Ty1_copia/Alesia:Ty1-RT 0.02% Class_I/LTR/Ty1_copia/Tork:Ty1-RH 0.02% Class_I/LTR/Ty1_copia/TAR:Ty1-PROT 0.02% Class_I/LTR/Ty1_copia/Bryco:Ty1-INT 0.02% Class_I/LTR/Ty1_copia/Alesia:Ty1-RH 0.02% Class_I/LTR/Ty1_copia/SIRE:Ty1-RT 0.02% Class_I/LTR/Ty1_copia/Angela:Ty1-INT |
| pair_completeness | 0.754950495049505 |
| pbs_score | None |
| TR_score | None |
| TR_monomer_length | None |
| loop_index | 0.66807710390221 |
| satellite_probability | 0.00559431351656918 |
| consensus | None |
| TAREAN_annotation | Other |
| orientation_score | 1 |
protein domains:

|
|
||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|